Searching for new ways to block the growth of cancer cells is an ongoing challenge in the field of cancer research. Tumor cells rely on numerous proteins to function properly, but identifying the specific proteins that can be targeted by drugs is a complex task. However, a recent study published in Nature Chemical Biology on October 2, 2023, by researchers at Scripps Research and the Broad Institute of Harvard and MIT has introduced a novel method to identify potential drug targets that could impact multiple types of cancer. This innovative approach integrates precise gene editing techniques with chemical proteomic data to narrow down the list of potential drug targets and discover new avenues for cancer treatment.
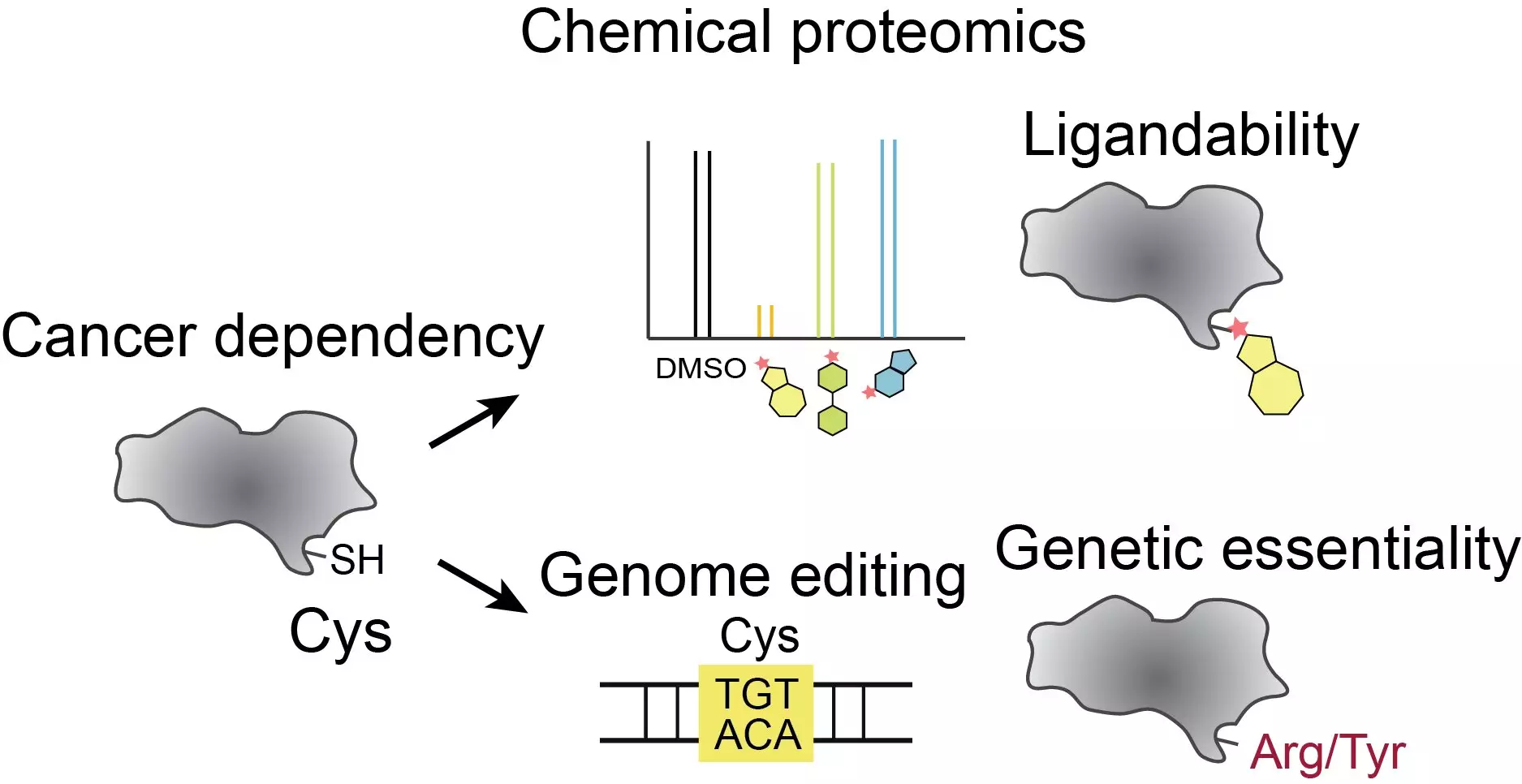
Over the past decade, researchers and pharmaceutical companies have shown great interest in drugs that permanently bind to cysteines, one of the amino acids present in human proteins. Cysteines have unique reactive properties that make them ideal drug targets. However, with hundreds of thousands of cysteines scattered throughout human proteins, it becomes challenging to pinpoint which cysteines to target with drugs. Even among the few thousand proteins known to be critical for cancer cell growth, there are still over 13,000 cysteines that could potentially be targeted.
In this study, the research team led by Benjamin Cravatt at Scripps Research and David Liu at the Broad Institute sought to narrow down the list of cysteines that have a significant functional impact on cancer-relevant proteins. To achieve this, they combined precise gene editing techniques with cutting-edge chemical proteomic tools.
The team edited more than 13,000 possible drug targets, specifically targeting cysteines on the corresponding proteins. They then assessed the impact of these edits on cancer cell growth. Additionally, the researchers integrated their findings with new data on the “druggability” of the cysteines. This integration of genomic and proteomic data allowed them to identify approximately 160 druggable cysteines that, when edited, significantly affected cancer cell growth. This discovery suggests that drugs binding to these cysteines could potentially be effective in treating cancer.
One of the most significant findings of the study was the identification of the cancer-dependency protein TOE1 as a potential drug target. While TOE1 is known for its role in trimming the ends of the cell’s RNA molecules, it had not been previously studied as a cancer drug target. However, the research team demonstrated that small molecules could be harnessed to target TOE1 and inhibit its normal activity, impairing cancer cell growth. This discovery highlights the value of this innovative approach in uncovering new drug targets that may have been overlooked in the past.
Although the initial results are promising, further research is needed to evaluate the effectiveness of drugs targeting TOE1 in human patients. Additionally, the researchers intend to explore other novel targets that emerged from their experiments. They also highlight the ongoing development of next-generation chemical genetic approaches to study druggable cysteines in diseases beyond cancer.
The study conducted by the team at Scripps Research and the Broad Institute presents an innovative method for identifying potential drug targets in cancer research. By combining precise gene editing techniques with chemical proteomic data, the researchers were able to narrow down the list of cysteines that have the greatest impact on cancer cell growth. This study lays the foundation for further exploration of new drug targets and highlights the potential of this approach in the field of cancer therapeutics.

Leave a Reply