Glycosylation, the attachment of carbohydrates to proteins through enzymatic reactions, is a critical process that greatly influences protein structure, function, and stability. Glycoproteins, the proteins that undergo glycosylation, play essential roles in various biological processes. There are two commonly known types of protein glycosylation: N-glycosylation and O-glycosylation. Researchers at the Boston University Chobanian & Avedisian School of Medicine recently conducted a study to enhance the identification of glycopeptides, molecules that contain one or more glycans, and their findings shed light on the importance of accurately identifying peptides with multiple glycans.
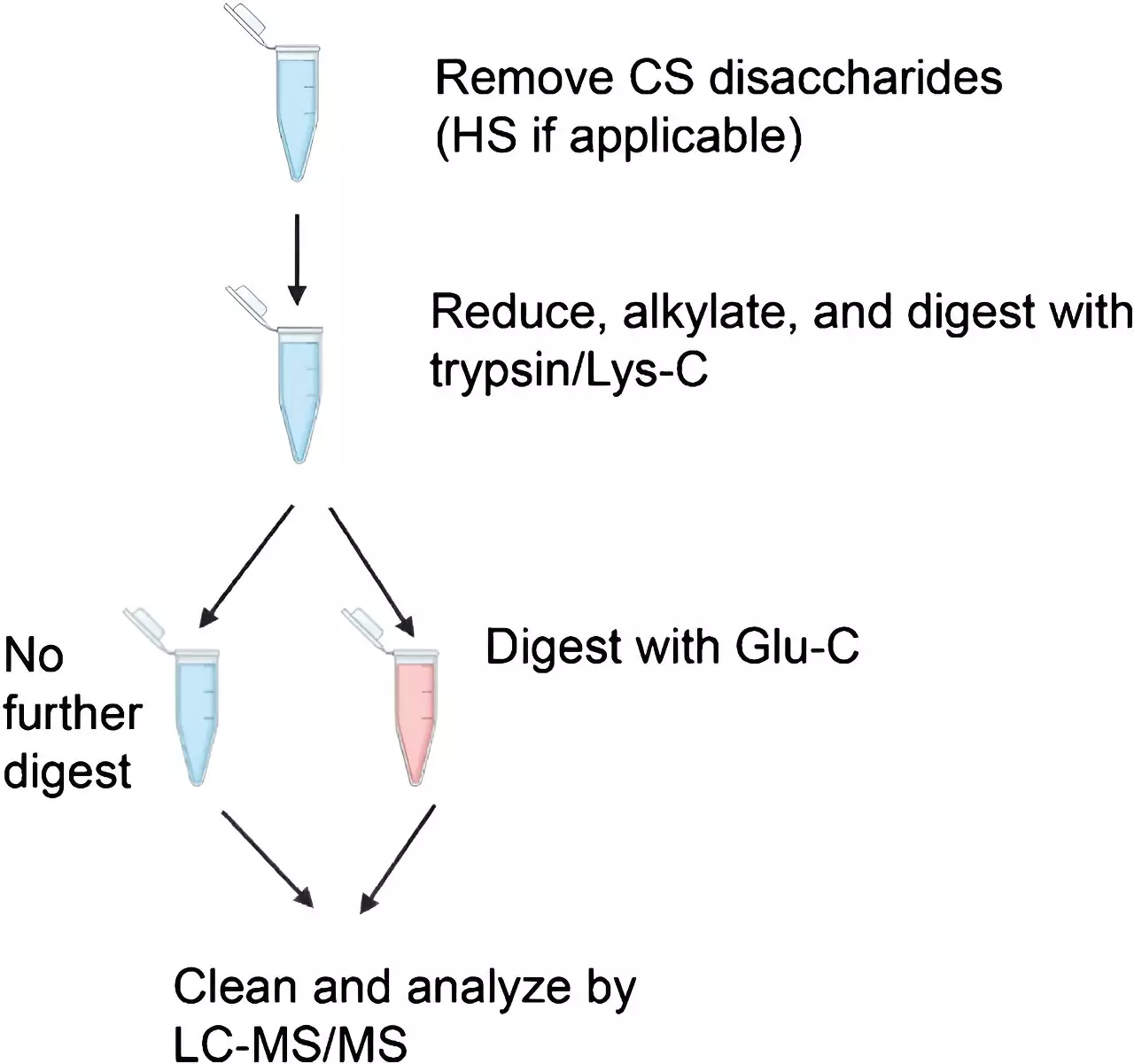
The study demonstrated that conventional mass spectrometry methods are effective in identifying peptides with one glycan. However, a crucial additional step is required to identify peptides with two or more glycans. Since a protein can carry numerous glycans, it is challenging to generate peptides containing only one glycan. Accurately identifying peptides with multiple glycans is vital for understanding the various glycoforms of a protein and comprehending its biological function and disease implications.
The lack of comprehensive information regarding glycoproteins and their changes during development and disease hinders the identification of promising therapeutic options. Manveen K. Sethi, the corresponding author of the study and a research assistant professor of biochemistry & cell biology, believes that a more specific understanding of how glycosylation affects disease, particularly site-specific alterations, could unveil novel therapeutic targets in diseases lacking effective treatment options. Sethi highlights that advances in technology enabling the confident identification of densely glycosylated peptides can aid in differentiating between healthy and diseased states based on site-specific glycosylation. This differentiation would facilitate the use of glycoproteins as markers and therapeutic targets in various diseases.
To validate their findings, the researchers utilized four different standard proteins and applied different enzymatic digestion protocols before conducting mass spectrometry analysis. They compared the conventional higher-energy collisional dissociation (HCD) method with a more recently developed method called electron transfer/higher energy collisional dissociation (EThcD). The aim was to determine the number of glycopeptides identified using each digestion protocol and mass spectrometry method. The results indicated that HCD is generally sufficient for glycopeptide identification. However, EThcD often becomes necessary, especially when identifying glycans in multiply-glycosylated peptides.
Glycosylation significantly influences protein structure, function, and stability, playing a critical role in various biological and disease processes. The ability to accurately identify peptides with multiple glycans is essential for understanding the diverse glycoforms of proteins and their implications in disease. The study conducted by the researchers at Boston University Chobanian & Avedisian School of Medicine highlighted the need for advances in technology that can confidently identify densely glycosylated peptides. This advancement would enable the differentiation between healthy and diseased states based on site-specific glycosylation, paving the way for the use of glycoproteins as markers and therapeutic targets in various diseases. With a deeper understanding of how glycosylation affects disease, researchers may discover novel and effective treatment avenues, addressing diseases that currently lack suitable therapeutic options.

Leave a Reply